Klap: The AI Video Editing Tool Revolutionizing Social Media Content Creation
-


Tired of apps that feel confusing or unreliable? Lotus365 cricket id brings...

Discover how IP rent services offer flexibility, security, and scalability...
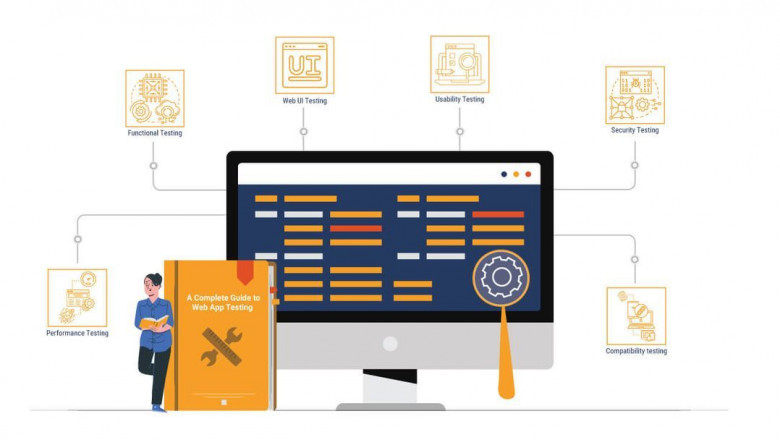
Anything is possible while deploying a website: broken links, web security,...

With people moving to a world more focused on digital products, software te...
Location: Qingdao, Shandong, China

The Sikkim Manipal University Online MCA is more than just a degree—it is a...

Looking for expert help? A top rated therapist in Santa Monica can guide yo...









